

The Genomics Core Laboratory in the Department of Pathology is established to house both basic and translational genomics research, allow individual investigators access to state-of-the-art next-generation sequencing (NGS) technologies, conduct genomics experiments and bioinformatics analyses, and provide opportunities for education and training in genomics and bioinformatics.
The Genomics Core Laboratory hosts a variety of instruments for carrying out genomics research. These instruments include, but are not limited to:
Qubit 2.0 Fluorometer for flurometric quantification of DNA, RNA and protein
Bioanalyzer 2100 for quantification, size and purity assessment of DNA, RNA and protein
TapeStation 4200 for quantification, size and purity assessment of DNA, RNA and protein
BluePippin for DNA size selection/collection ranged from 100 bp to 50 kb with pulse-field
Stratagene Mx3005P for real-time quantitative PCR
SpectraMax Gemini XPS microplate reader for fluorescence intensity assays
Biomek FXp automated workstation for high-throughput liquid handling and workflow
MiSeq for single- or paired-end short-read sequencing ranged from 50 bp to 300 bp
NextSeq 550 for single- or paired-end short-read sequencing ranged from 50 bp to 150 bp
Please click the instrument name for the link to its official website for overview and/or introduction.
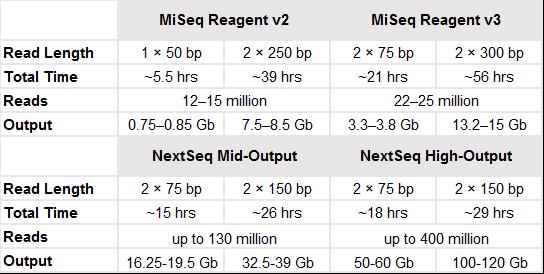
The Genomics Core Laboratory mainly uses the Illumina MiSeq and NextSeq two sequencing systems as the NGS platform. The specifications of MiSeq and NextSeq are demonstrated in the table below.
The Genomics Core Laboratory provides and supports a variety of NGS services and their following bioinformatics data analyses. Consultation before services is freely available and is strongly recommended.
The NGS services include:
The bioinformatics services include:
Core Pricing:
Consultation for the total cost of planned service is thus strongly recommended. Since the bioinformatics services are free of charge, a courtesy from the Genomics Core Laboratory, the co-authorship in your publications is greatly appreciated.
| NA1:E58extSeq 550 | ||||
| Service | Internal | External | ||
| Mid-Output | ||||
| 150 bp sequencing | $1297.00 | $1362.00 | ||
| (2x75 bp) | ||||
| Mid-Output | ||||
| 300 bp sequencing | $2090.00 | $2195.00 | ||
| (2x150 bp) | ||||
| High-Output | ||||
| 150 bp sequencing | $3410.00 | $3581.00 | ||
| (2x75 bp) | ||||
| High-Output | ||||
| 300 bp sequencing | $5462.00 | $5735.00 | ||
| (2x150 bp) | ||||
| MiSeq | ||||
| Service | Internal | External | ||
| V2, 50 bp sequencing | ||||
| (1x50 bp) | $980.00 | $1029.00 | ||
| V2, 500 bp sequencing | ||||
| (2x250 bp) | $1410.00 | $1481.00 | ||
| V3, 150 bp sequencing | ||||
| (2x75 bp) | $1088.00 | $1142.00 | ||
| V3, 600 bp sequencing | ||||
| (2x300 bp) | $1841.00 | $1933.00 | ||
| Library Prep | ||||
| Service | Internal | External | ||
| TruSeq mRNA | ||||
| (polyT purification) | $ 160.00 | $ 192.00 | ||
| TruSeq mRNA | ||||
| (RiboZero purification) | $ 275.00 | $ 330.00 | ||
| TruSeq small RNA | $ 300.00 | $ 360.00 | ||
| Nextera XT | $ 100.00 | $ 120.00 | ||
| Others | ||||
| Service | Internal | External | ||
| Bioanalyzer 2100 | ||||
| (DNA or RNA, per sample) | $ 15.00 | $ 20.00 | ||
| TapStation 4200 | ||||
| (DNA or RNA, per sample) | $ 12.00 | $ 15.00 | ||
| BluePippin | ||||
| (size selection, per sample) | $ 25.00 | $ 30.00 | ||
| Stratagene Mx3005p | ||||
| Real-time PCR | inquire | inquire | ||
| SpectraMax Gemini XPS | ||||
| Fluorometer 96-well | inquire | inquire | ||
| Biomek FXp | ||||
| (automation system) | inquire | inquire | ||
Genomics Core Laboratory is located at Basic Sciences Building Room 409 at Pathology Department. Investigators are welcome to visit, consult and request services of genomics and bioinformatics.
Humayun Islam, M.D., Ph.D.
Chair of Pathology, Microbiology and Immunology
Phone: (914) 594-4150
Fax: (914) 594-4163
Humayun.Islam@wmchealth.org